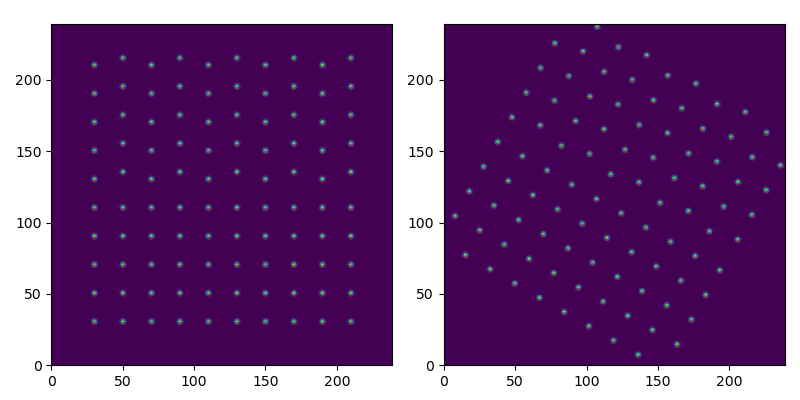
Various tools
Rotating image and points
>>> from scipy.ndimage import rotate
>>> import atomap.api as am
>>> from atomap.tools import rotate_points_around_signal_centre
>>> import matplotlib.pyplot as plt
>>> sublattice = am.dummy_data.get_distorted_cubic_sublattice()
>>> s = sublattice.get_atom_list_on_image()
>>> s_orig = s.deepcopy()
>>> rotation = 30
>>> s.map(rotate, angle=rotation, reshape=False)
>>> x, y = sublattice.x_position, sublattice.y_position
>>> x_rot, y_rot = rotate_points_around_signal_centre(s, x, y, rotation)
>>> fig, (ax0, ax1) = plt.subplots(1, 2, figsize=(8, 4))
>>> cax0 = ax0.imshow(s_orig.data, origin='lower', extent=s_orig.axes_manager.signal_extent)
>>> cax01 = ax0.scatter(x, y, s=0.2, color='red')
>>> cax1 = ax1.imshow(s.data, origin='lower', extent=s.axes_manager.signal_extent)
>>> cax11 = ax1.scatter(x_rot, y_rot, s=0.2, color='red')
>>> a = ax1.set_xlim(s.axes_manager[0].low_value, s.axes_manager[0].high_value)
>>> b = ax1.set_ylim(s.axes_manager[1].low_value, s.axes_manager[1].high_value)
>>> fig.tight_layout()
>>> fig.savefig("rotate_image_and_points.png", dpi=200)
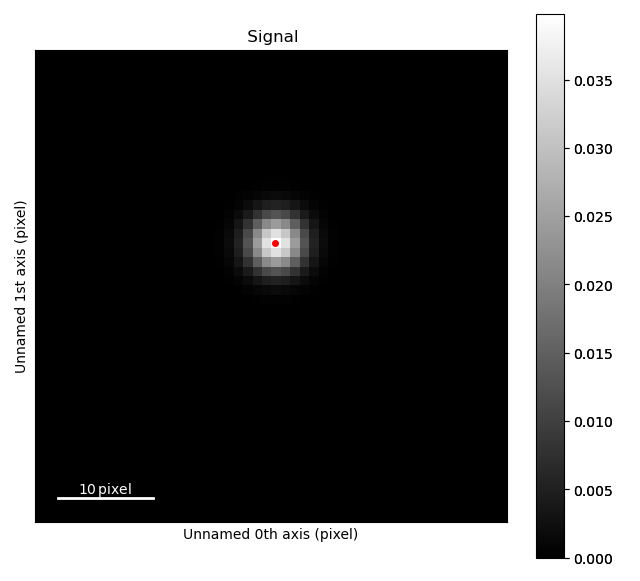
Working with single atom sublattices
Normally refining the atom positions requires using knowing the distances to the
atom’s nearest neighbors, to avoid fitting overlap.
However, with sublattices consisting of a single atom, this clearly does not work.
The mask_radius argument in refine_atom_positions_using_center_of_mass()
and refine_atom_positions_using_2d_gaussian() can be used in this case.
>>> sublattice = am.dummy_data.get_single_atom_sublattice()
>>> sublattice.refine_atom_positions_using_center_of_mass(mask_radius=9)
>>> sublattice.refine_atom_positions_using_2d_gaussian(mask_radius=9)
>>> sublattice.plot()